About
The Cardiac Epigenome Browser is a resource for cell-type specific cardiac chromatin interaction and epigenome data. The project is located at University of Freiburg (Institute of experimental and clinical Pharmacology and Toxicology).
Background
The heart is composed of different cell-types including cardiac myocytes, fibroblasts and endothelial cells. Their relative portion depends on disease and developmental stage. Gene regulation cells mainly depends on epigenetic mechanisms. Due to the highly cell-type specific nature of these mechanisms analysis of pure cell-types is essential. Cardiac myocyte-specific epigenetic data can i.e. be obtained from FACS-sorted cardiac myocytes or cardiac myocyte nuclei (Figure, Gilsbach et al. 2014; Preissl et al. 2015).
Within the restricted nuclear space, the DNA is compacted together with proteins (chromatin) in a highly structured manner. Chromatin interaction analysis by Hi-C-based methods gives insights into the hierarchically organization of the chromatin. This enables identification of interacting chromatin domains (TADs), active and inactive compartments (A/B compartments) and finally chromatin contact points like promoter enhancer contacts. These potential regulatory regions can further be characterized using specific chromatin marks and DNA methylation signatures.
The Cardiac Epigenome Browser allows visualization of chromatin interaction data together with various chromatin marks, DNA methylation and gene expression.
Usage
The Cardiac Epigenome Browser is based on the HiCBrowser. Select the organism in the menu and directly access cardiac myocyte specific data. Just type in your favorite gene name or a chromosomal region in the search field and the data is visualized.
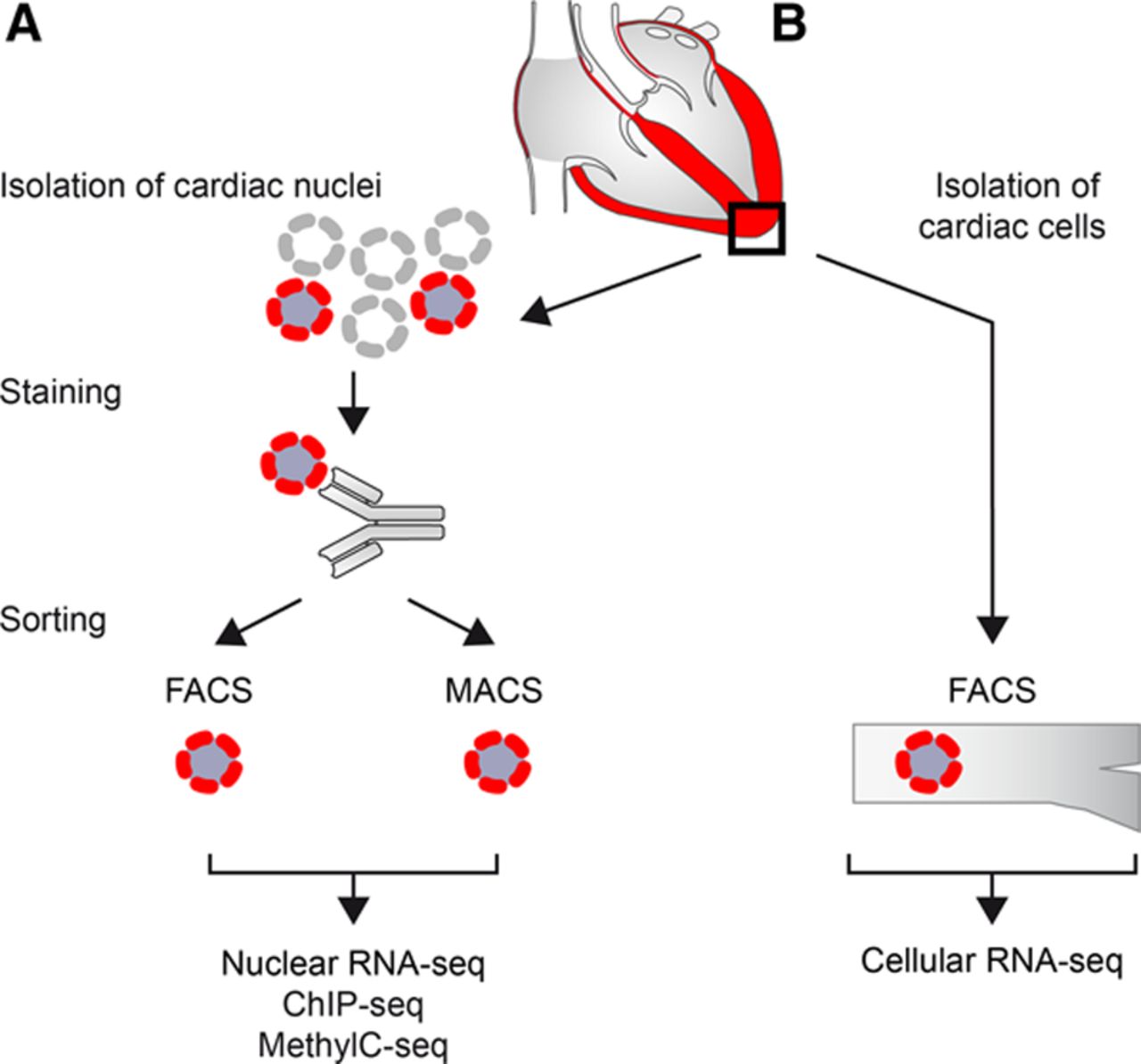
Figure: Isolation of cardiac nuclei or cells (from Preissl et al. 2015)

